pTAx vectors are built from five PCR products. This Google sheet contain primer numbers and template plasmids and PCR target for each of the five PCR products for all current pTAx vectors. Each PCR reaction is marked with a different color. All further information needed to assemble a pTAx vector in-silico (sequences, etc.) is available on this page. Table 1 contain plasmid sequences and Table 2 all primer sequences (below).
Each of the five PCR products is amplified using primers with tails (Figure 1), incorporating 30 bp flanking sequences ❶ and ❷.
Figure 1
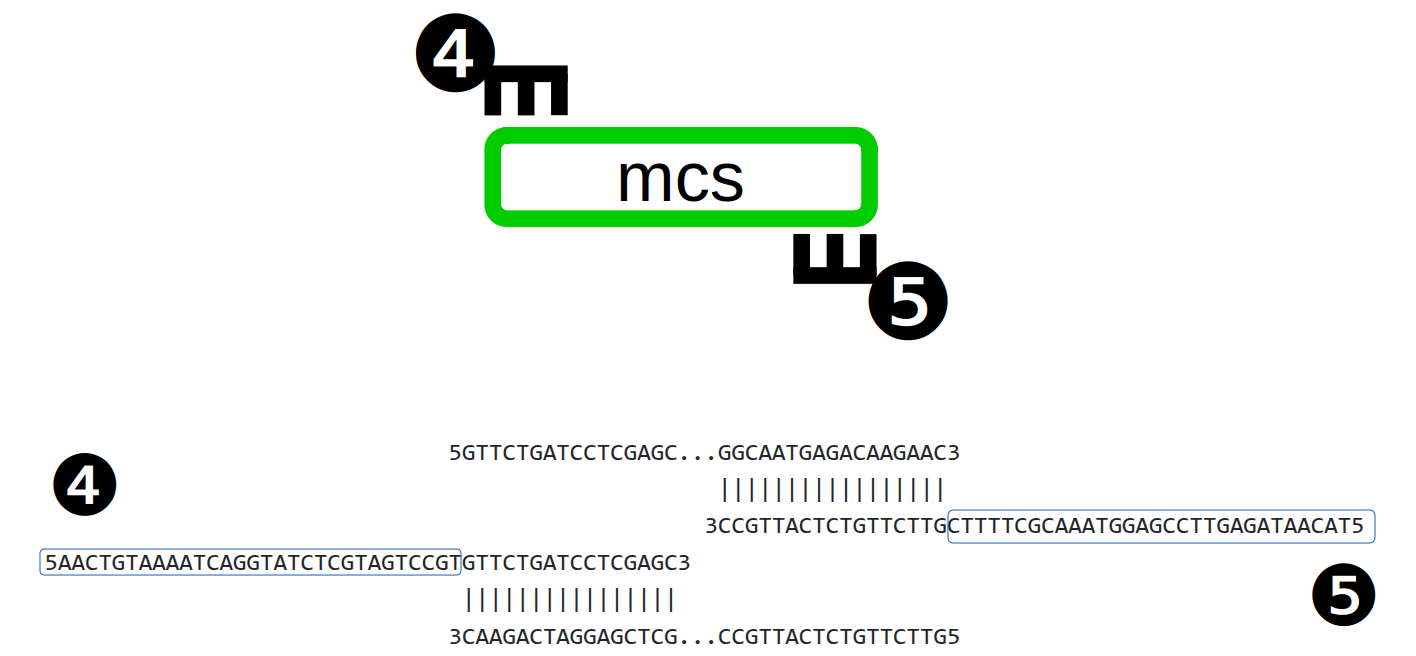
Five different specific flanking sequences are added to the end of each of the PCR products, (❶..❺) in Figure 2.
Figure 2

These flanking sequences enable homologous recombination between the DNA fragments in a specific order. The vectors are assembled by homologous recombination between the five sequences (❶..❺) as shown in Figure 3.
Figure 3
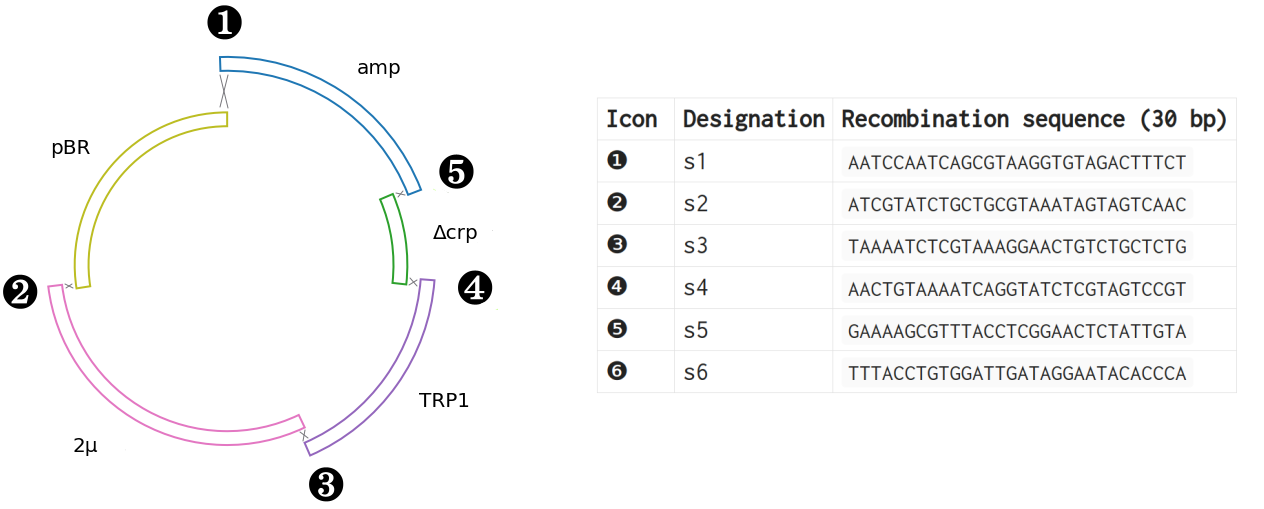
The plasmids can be assembled using the Pydnaweb PCR and assembly simulators. Use the links in the Recommended sequence column in Table 1 and the primer sequences in Table 2. Fragments are joined by homologous recombination between the 30 bp flanking DNA sequences (s1-s5; table).
Table 1 Plasmid links
Links to sequences and references for each source plasmid.
| Table1 | Plasmid | Freezer# | Reference | Genbank (Gb) | Snapgene (Sg) | Gb vs. Sg | 👌 Recommended sequence | seguid checksum |
|---|---|---|---|---|---|---|---|---|
| pBR322 | µ831 | link | J01749 | link | Same size; 1 nt missense in Tc gene. | Gb | cdseguid=H-FY2ZzvKeazrRW2dNeSeMikjoc | |
| pTA1 | µ828, µ928, µ929 | link | - | - | GitHub | cdseguid=MtltqcR0b0kVX_FJ4BYL-PjEAfg | ||
| pYPKpw | µ463 | link | - | - | GitHub | cdseguid=xj7evEO4h83-RGFeHu8q15tL_Qs | ||
| p423GPD | µ601 | link | - | - | GitHub | |||
| pFA6a-GFPS65T-kanMX6 | µ716 | link | AJ002682.1 | link | cdseguid=fisQRzPkD-XD07m2YQFOa2B1NdA | |||
| pSH65 | µ853 | link | AF298780.1 (partial) | - | Genbank has a 1 nt insertion. | GitHub | cdseguid=FEU5H5BV4yVTVCANkejX4c3kOVk | |
| pUG35 | µ762 | - | AF298787.1 | - | cdseguid=4cVPUTb2zz_rtTp5F2mci2l09rQ | |||
| YEplac112 | µ661 | link | X75458.1 | link | 1 snp in pUC origin | GitHub | cdseguid=MLKxSGFkZsuuiqozB87sVQQ0xaU | |
| YEplac195 | µ662 | link | X75459.1 | link | 1 snp in pUC origin | GitHub | cdseguid=nsxZh92qaUyjufyG0goiB3-Aq8c | |
| YEplac181 | µ663 | link | X75460.1 | link | 12 bp deletion in end of LEU2 promoter in Gb. | GitHub | cdseguid=gZjQlzVQtwNzfV1XOFu1psiO2Eo | |
| YIplac204 | µ664 | link | X75461.1 | link | 1 snp in pUC origin | GitHub | cdseguid=24fcVzVrHWTQXwerHr3_OyEw9D0 | |
| YIplac211 | µ665 | link | X75462.1 | link | 1 snp in pUC origin | GitHub | cdseguid=U-L4v2SU5qf0bIra0ANlXaGVXlM | |
| YIplac128 | µ666 | link | X75463.1 | link | Deletion in start of LEU2 for Gb | GitHub | cdseguid=Ch6Y7zxAoVN5PAGDHKdzT2AFmm8 | |
| YCplac22 | µ658 | link | X75455.1 | link | GitHub | cdseguid=BB-iNIXT2u55EuDhEHH0yufrGWg | ||
| YCplac33 | µ659 | link | X75456.1 | link | GitHub | cdseguid=XyTO5X31FUQgGt9m4roqPDTDzZs | ||
| YCplac111 | µ660 | link | X75457.1 | link | GitHub | cdseguid=7ytS0ma8bAjM9lEcozGnQDYtse8 |
Table 2 Primer sequences
>1750_s1 s1 tm=52.856
AATCCAATCAGCGTAAG
>1749_s2 s2 tm=56.264
ATCGTATCTGCTGCGT
>1748_s3 s3 tm=53.243
TAAAATCTCGTAAAGGAACT
>1747_s4 s4 tm=52.983
AACTGTAAAATCAGGTATCT
>1746_s5 s5 tm=54.852
GAAAAGCGTTTACCTCG
>1745_s1r s1r tm=52.348
AGAAAGTCTACACCTTAC
>1744_s2r s2r tm=54.01
GTTGACTACTATTTACGCA
>1743_s3r s3r tm=55.045
CAGAGCAGACAGTTCC
>1742_s4r s4r tm=53.771
ACGGACTACGAGATAC
>1741_s5r s5r tm=51.462
TACAATAGAGTTCCGAG
>1222_2954fw 29-mer
GAAATTTCAGCCACTTCACAGGCGGTTTT
>1779_pUCmu_bb_R 24-mer
actcttcctttttcaatattattg
>1804_TRP1fp_pTA replaces 1348 & 1680
TAAAATCTCGTAAAGGAACTGTCTGCTCTGtataaaaataggcgtatcacg
>1691_URA3fp_pTA_2
TAAAATCTCGTAAAGGAACTGTCTGCTCTGgcttttcaattcaattcatc
>1690_HIS3rp_pTA_2
ACGGACTACGAGATACCTGATTTTACAGTTgacacgtatagaatgatg
>1681_Pbr.FW.2 New pBR primer, partial s1, replaces 1196
TAAGGTGTAGACTTTCTccgtagaaaagatcaaag
>1680_TRP1fp_pTA New TRP1 primer, partial s3, replaces 1348, replaced by 1804
GAACTGTCTGCTCTGttcaagaattaattcggtcg
>1352_HIS3fp_pTA
TAAAATCTCGTAAAGGAACTGTCTGCTCTGAACACAGTCCTTTCC
>1351_HIS3rp_pTA
ACGGACTACGAGATACCTGATTTTACAGTTTTTTTTCTCGAGTTCAAGA
>1350_KanMX4fp_pTA
TAAAATCTCGTAAAGGAACTGTCTGCTCTGGACATGGAGGCCC
>1349_KanMX4rp_pTA
ACGGACTACGAGATACCTGATTTTACAGTTCAGTATAGCGACCAGC
>1348_TRP1fp_pTA
TAAAATCTCGTAAAGGAACTGTCTGCTCTGTTTGCCGATTAAGAATTCG
>1347_TRP1rp_pTA
ACGGACTACGAGATACCTGATTTTACAGTTGATCTTTTATGCTTGCTTTTC
>1346_URA3fp_pTA
TAAAATCTCGTAAAGGAACTGTCTGCTCTGGATTCCGGTTTCTTTGAAA
>1345_URA3rp_pTA
ACGGACTACGAGATACCTGATTTTACAGTTGGTAATAACTGATATAATTAAATTGAA
>1344_CEN_ARSfp_pTA
ATCGTATCTGCTGCGTAAATAGTAGTCAACACGGATCGCTTGC
>1343_CEN_ARSrp_pTA
CAGAGCAGACAGTTCCTTTACGAGATTTTACTTTTCATCACGTGCTATA
>1196_Pbr.FW
AATCCAATCAGCGTAAGGTGTAGACTTTCTCTGTCAGACCAAGTTTACT
>1195_Pbr.REV
GTTGACTACTATTTACGCAGCAGATACGATCTCGTTTCATCGGT
>1113_Amp.fw.nw
GAAAAGCGTTTACCTCGGAACTCTATTGTAGAACCCCTATTTGTTTATTTTTCTA
>988_Amp.FW Amp.FW (do not use!)
GAAAAGCGTTTACCTCGGAACTCTATTGTAAACCCTGATAAATGCTTC
>987_Amp.REV Amp.REV
AGAAAGTCTACACCTTACGCTGATTGGATTTGAAGTTTTAAATCAATCTAAA
>986_Pbr.FW Pbr.FW
AATCCAATCAGCGTAAGGTGTAGACTTTCTCTCGTTTCATCGGTAT
>985_Pbr.REV Pbr.REV
GTTGACTACTATTTACGCAGCAGATACGATCTGTCAGACCAAGTTTACT
>984_Micron.FW Micron.FW
ATCGTATCTGCTGCGTAAATAGTAGTCAACGATCGTACTTGTTACCCAT
>983_Micron.REV Micron.REV
CAGAGCAGACAGTTCCTTTACGAGATTTTAGATCCAATATCAAAGGAA
>982_dLeu.FW dLeu.FW
TAAAATCTCGTAAAGGAACTGTCTGCTCTTATATATATTTCAAGGATATACCATT
>981_dLeu.REV dLeu.REV
ACGGACTACGAGATACCTGATTTTACAGTTTAAGGCCGTTTCTGAC
>980_Leu2.FW Leu2.FW
TAAAATCTCGTAAAGGAACTGTCTGCTCTGTTAACTGTGGGAATACTC
>979_Leu2.REV Leu2.REV
ACGGACTACGAGATACCTGATTTTACAGTTTCTACCCTATGAACATATTC
>978_Crp.FW Crp.FW
AACTGTAAAATCAGGTATCTCGTAGTCCGTGTTCTGATCCTCGAGC
>977_Crp.REV Crp.REV
TACAATAGAGTTCCGAGGTAAACGCTTTTCGTTCTTGTCTCATTGCC