In-vivo DNA assembly by homologous recombination in Saccharomyces cerevisiae via single-strand annealing
The figure below illustrates how linear DNA fragments can be joined inside the budding yeast S. cerevisiae using its natural homologous recombination machinery, a process known as “gap-repair”. Homologous recombination most likely happens though the Single Strand Annealing (SSA) mechanism:
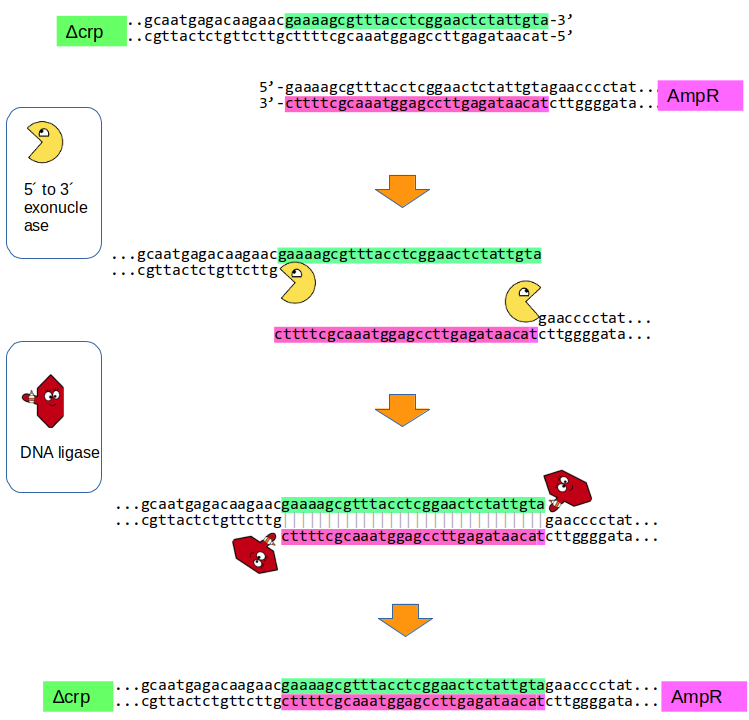
The DNA fragments share short regions of sequence homology (about 20-30 base pairs) at their ends. 5′ to 3′ exonucleases inside the yeast cell (The yellow pac-man) activity removes nucleotides from the ends of the DNA, generating single-stranded 3′ overhangs (sticky-ends). These overhangs allow the homologous regions to anneal to each other.
The annealing step is thought to occur through alternative-end joining (alt-EJ) or single-strand annealing (SSA), two DNA repair pathways active in yeast that do not require complex recombination machinery. After annealing, any gaps are filled in by yeast DNA polymerases and the nicks are sealed by DNA ligase (The red hexagon with the glue), resulting in the formation of a continuous DNA fragment.
See Vu, Tien Van, Swati Das, Cam Chau Nguyen, Jihae Kim, and Jae-Yean Kim. 2022. “Single-Strand Annealing: Molecular Mechanisms and Potential Applications in CRISPR-Cas-Based Precision Genome Editing.” Biotechnology Journal 17 (7): 2100413. https://doi.org/10.1002/biot.202100413.