Primer design and melting temperature calculations for PCR primers
In TP04 we learned how to design primers given a template sequence. This exercise will show you how to design PCR primers that can work well in a real PCR.
A typical PCR program is shown below. It is important to adjust the annealing temperature (50 ºC in the example below). This annealing temperature depends mostly on the PCR primers.
Each primer has a melting temperature (Tm) which depend mostly on the GC content and length of the primer. Primer melting temperature (Tm) by definition is the temperature at which one half of the DNA duplex will dissociate to become single stranded and indicates the duplex stability.
A common strategy is to use the lowest primer melting temperature as the annealing temperature (Ta), although more elaborate algorithms are also used.
| 30 cycles |
| |
|denaturation| anneal |extension |
| |
95C | 95C |
_____|___________ 72C | 72C
4min | 30s \ __________|_______
| \ 50C / 1min30s | 4min
| \_____/ |
| 30s |
We need the Tm for both primers in order to estimate the Ta of the PCR reaction. The simplest melting temperature formula that can be used is called the Basic formula also sometimes called the Marmur formula:
Tm = (A+T) * 2 + (G+C) * 4
Where A, T, G and C are the number of the respective bases in the sequence (modified from Marmur, J., and Doty, P. 1962 J Mol Biol 5:109-118). According to this formula, the Tm for the primer below is 38ºC:
>primer
gatcatctcgatc
Question 1:
What is the Tm of primer2 below using the Basic/Marmur formula?
>primer2
cacttcctgacatcg
DNA melting
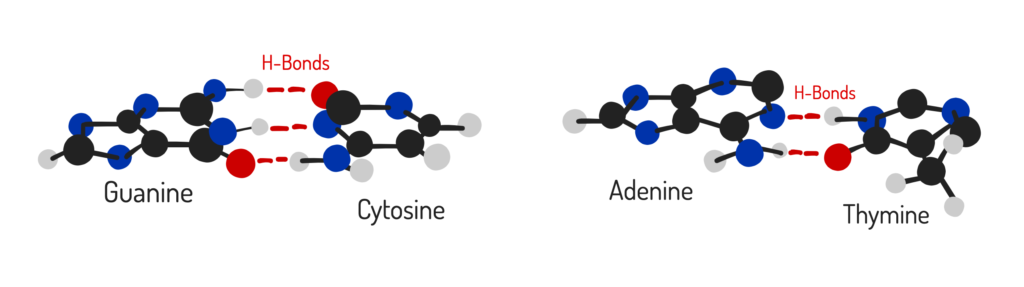 Hydrogen Bonds: While hydrogen bonds (G≡C has 3, A=T has 2) contribute, base stacking – the attraction between the flat surfaces of adjacent bases in the helix – is a major determinant of stability.
Hydrogen Bonds: While hydrogen bonds (G≡C has 3, A=T has 2) contribute, base stacking – the attraction between the flat surfaces of adjacent bases in the helix – is a major determinant of stability.
GC vs. AT Stacking: GC-rich DNA has much stronger stacking interactions (e.g., GC-GC stacks are significantly more stable than AT-AT stacks).
There are also more elaborate formulas for Tm calculation. These are often implemented in special software tools, many available online. Many tools are based on nearest neighbor calculations of the melting temperature.

The nearest-neighbor (NN) model for nucleic acids predicts DNA thermodynamics using energy values for the different base pair groups. These values have been derived from melting experiments in monovalent and divalent salt. The YouTube video above explaining the Nearest Neighbour algorithm_
Unfortunately, most tools give very different results. The reason for this is not always obvious and most tools are not sufficiently documented to find out.
One of the best documented Tm algorithms available is the one used in Biopython. Biopython is an open source Python software package focused on bioinformatics. This algorithm is used in the Pydna Tm calculator.
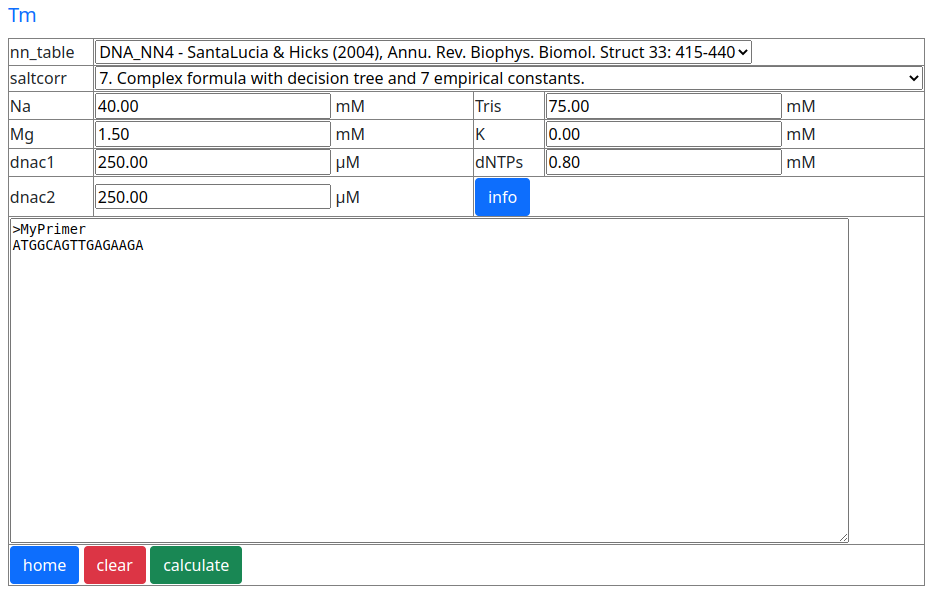
Question 2:
What is the Tm of the oligonucleotide cacttcctgacatcg using the default settings of the Pydnaweb Tm tool (above)?
Question 3:
This is an individual exercise for each student. The input data can be found in a the TP05 Google spreadsheet where you can find your name in the leftmost column.
One column called geneY contains a DNA sequence that represents a double stranded linear DNA molecule that is also an open reading frame. Your task is to design a reverse primer (rp) for geneY that will amplify all of the sequence together with the given forward primer (fp). The sequence of fp is given in the spreadsheet.
The melting temperature of rp should match the one of fp as closely as possible using the Pydnaweb Tm calculator with the default settings.
Put your results in the indicated cells reverse primer (rp). Please answer with raw DNA sequences as indicated for the first example student “Max Maximus”.
© Björn Johansson 2013-2026